ON EVOLUTIONARY QUANTIFICATIONS OF STRONGLY DISTORTED MR-SIGNALS
Ales Gottvald
Institute of Scientific Instruments, Academy of Sciences of the CR,
Kralovopolska 147, CZ-612 64 Brno, Czech Republic
E-mail: gott@isibrno.cz , Internet: http://www.isibrno.cz/~gott/index.html
Abstract: Non-Fourier quantification methodologies show some principal advantages for quantifying strongly
distorted MR-signals [1 - 6]. A very general methodology of quantitative MRS may be based on Meta-Evolutionary
optimization and Bayesian Statistics [6]. This robust probabilistic approach has been applied to a class of
generalized FID-signals, developed for inhomogeneity, noise, phase, truncation, apodization, and some other
artifacts.
From a mathematical point of view, we are facing to a typical inverse problem (ill-posed problems in the sense
of Hadamard). Associated optimization formulation may be stated as follows:
minimize: f(x):= | u* - u(x) |, subject to: x e X c Rn (1)
where u* is the input data-vector (a sampled FID-signal, experimental or synthetic), x e Rn are the parameters to
be determined, u(x) is a parametric model (in our case - associated with a generalized FID-signal), |.| is a norm,
and a configuration manifold X is defined by a set of constraints (equations and inequalities).
Uncertainty of solutions is an inherent feature of inverse problems. For every inverse problem, there exist some
natural physical limits for the accuracy of parameter estimates. These fundamental statistical limits (e.g., Cramer-Rao
lower bounds) cannot be superseded in any way without using some additional prior information. Consequently, the
major "secret" of solving the inverse problems consists in identifying and applying all forms of prior information -
both hidden and explicit. It may be shown [6] that important prior information is associated with all structural
elements in (1), including the noise-associated norm |.|, constraints defining x e Rn, etc. For strongly distorted
signals, an important prior information is involved in the parametric model itself (see accompanying two
abstracts). In our approach, inhomogeneity-asociated prior information is neither "guessed" nor "neglected", but
identified using experimental data. This identification operation actually reduces a number of degrees of freedom
to be determined via the optimization or inversion (1).
Meta-Evolutionary optimization [6] is a stochastic optimization method inspired by a biological evolution of
species. In contrast to standard stochastic optimizations (Evolution Strategy, Simulated Annealing, Genetic
Algorithms), this meta-method provides some additional quarantees: enforced global convergence, reduced
sensitivity to initial parameter estimates, and detailed uncertainty analysis (inversion errors, total accuracy estimates,
fundamental statistical limits). When being linked to a Bayesian formulation, noise features (and some probabilistic
prior information) may be respected. For example, an additive Gaussian white noise in the data may be related to
a Maximum Likelihood Estimation using L2-norm. A Bayesian Statistic is than generated by sampling an associated
error function f(x) with the Meta-Evolutionary optimizer.
An example of the evolutionary quantification, for a 31P In Vivo MR-spectrum of a human's brain, is indicated
in Fig. 1. In this case, a synthetic spectrum with strong inhomogeneity and noise artifacts has been optimally restored,
in terms of the most likely estimates for 21 spectral parameters. The difference between the conventional DFT-based
spectrum (Fig. 1b) and its optimally reconstructed version (Fig. 1d) is remarkable. It illustrates that even a rough
specification of prior information is clearly preferable to its full negligence or an ad hoc choice (as typical for a
conventional DFT-based quantification).
References:
[1] de Beer R. - van Ormondt D.: "Analysis of NMR Data Using Time Domain Fitting Procedures". NMR - Basic Concepts and Progress,
Springer-Verlag, Berlin, 1992, pp. 202 - 248
[2] Haselgrove J. C. et al.: "Analysis of In Vivo NMR Spectra". Reviews of Magn. Reson. in Medicine 2, 2, 1987, pp. 167 - 222
[3] Bretthorst G. L.: "Bayesian Analysis. ...", J. Magn. Reson. 88 (1990), 533 - 551
[4] Kotyk J. J. et al.: "Comparison of Fourier and Bayesian Analysis of NMR Signals". Part I. - J. Magn. Reson. 98, 1992, pp. 483 - 500; Part
II. - J. Magn Reson. A 116, 1995, pp. 1 - 9
[5] Johnson G. et al.: "Multiple-Window Spectrum Estimation Applied to in Vivo NMR Spectroscopy". J. Magn. Reson. B 110, 1996, pp. 138 -
149
[6] Gottvald A.: "A Survey of Inverse Methodologies, Meta-Evolutionary Optimization and Bayesian Statistics: Applications to In Vivo MRS".
Intnl. J. of Appl. Electromagnetics and Mechanics, 1996/97 (accepted)
Acknowledgment: This work was supported in part by GA CR, grant # 102/95/0282.
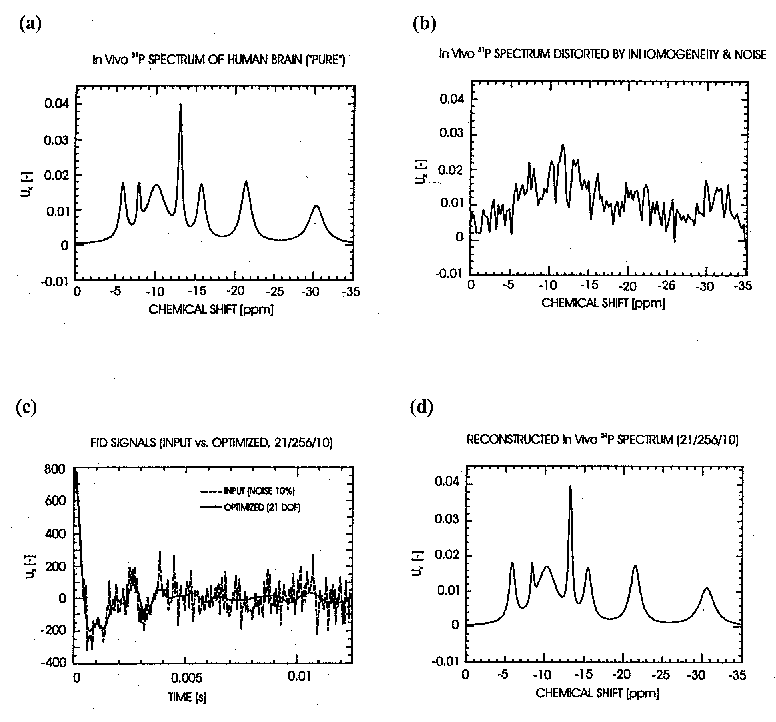
Fig. 1: The most likely reconstruction of a synthetic In Vivo 31P-spectrum (real part) with strong inhomogeneity and
noise distortions. (a) An ideal "pure" spectrum. (b) Standard DFT-spectrum with inhomogeneity and noise
distortions, without apodization. (c) Time-domain signals - input noisy data and reconstructed optimal signal. (d)
Reconstructed spectrum - inhomogeneity and noise distortions eliminated. Conditions: 21 spectral parameters
determined via Meta-Evolutionary Bayesian Statistics, imposing only insignificant soft constraints on resonant
frequencies, amplitudes and phases (relaxation rates fixed). Real parts of the signal and the spectra are shown.
Number of complex data-values: 256. Noise: white Gaussian, 10 % of the maximal signal amplitude.
Synthetic FID-signals generated from experimental parameters given in [2], using an ad hoc inhomogeneity function.
For 20 Meta-optimization loops, this inversion requires CPU-time ~ 100 seconds/1 MFLOPS.
 Back to:
Back to:

 Back to:
Back to: